Service Charges and Ordering
Service prices
The Core offers 3’-end gene expression profiling of up to 10,000 cells per sample. Full service from cell capture, cDNA synthesis, subsequent library preparation, Illumina sequencing to data analysis by DRAGEN are available. Please contact Dr. KWOK, Hin (pipsinglecell.cpos@hku.hk / 2831-5483) for project discussion and service quotation.
Prices vary with the following factors:
- Number of target cells (affecting the sequencing cost)
- Bioinformatics analysis needs
- Affiliated institute
Service ordering
Please refer to Sample Requirement and Submission section for sample requirements.
Please submit service request through iLab.
Technical Details
Cell Capture and Library Preparation
Cells in suspension are first mixed with barcoded hydrogel template particles and oil. By vortexing, cells are instantaneously captured in particle-templated instant partitions (PIPs). They are then lysed to release their mRNA, which are in turn captured by poly(dT) primers on the barcoded beads. Subsequently, the emulsion is broken and washed to recover the beads for reverse transcription.
Next, full length cDNA is fragmented, ligated with adapters and amplified into libraries in bulk. Final libraries will contain unique cell barcodes and intrinsic molecular identifiers (IMIs) for more accurate cell and molecular counting.
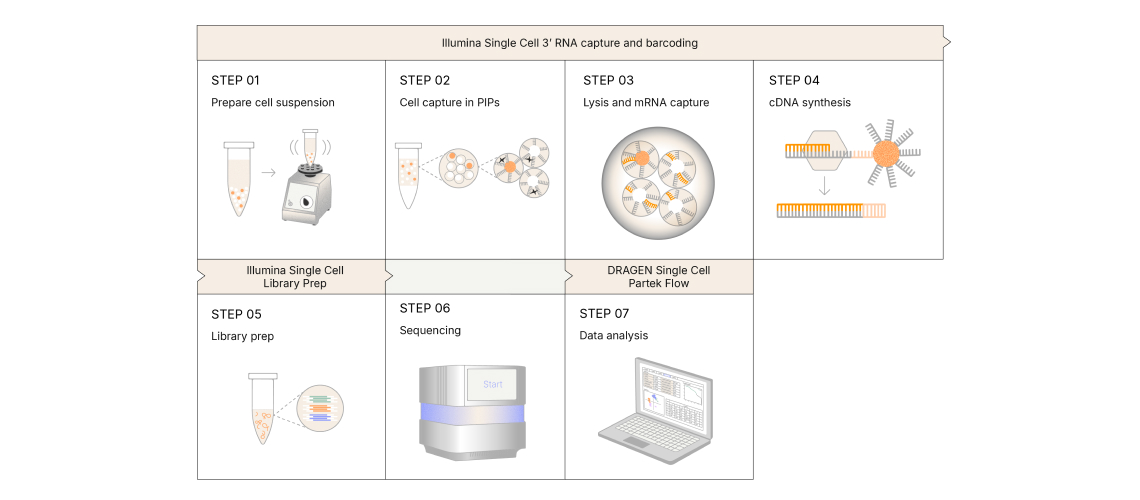
Source: Illumina
Sequencing Run
Libraries generated are compatible with Illumina sequencers. In order to read the transcript sequences on one end, and the barcode on the other end, paired-end sequencing reads are required. Sequencing depth varies based on user application needs but vendor recommends to start with a depth of 20,000 read pairs per input cell.
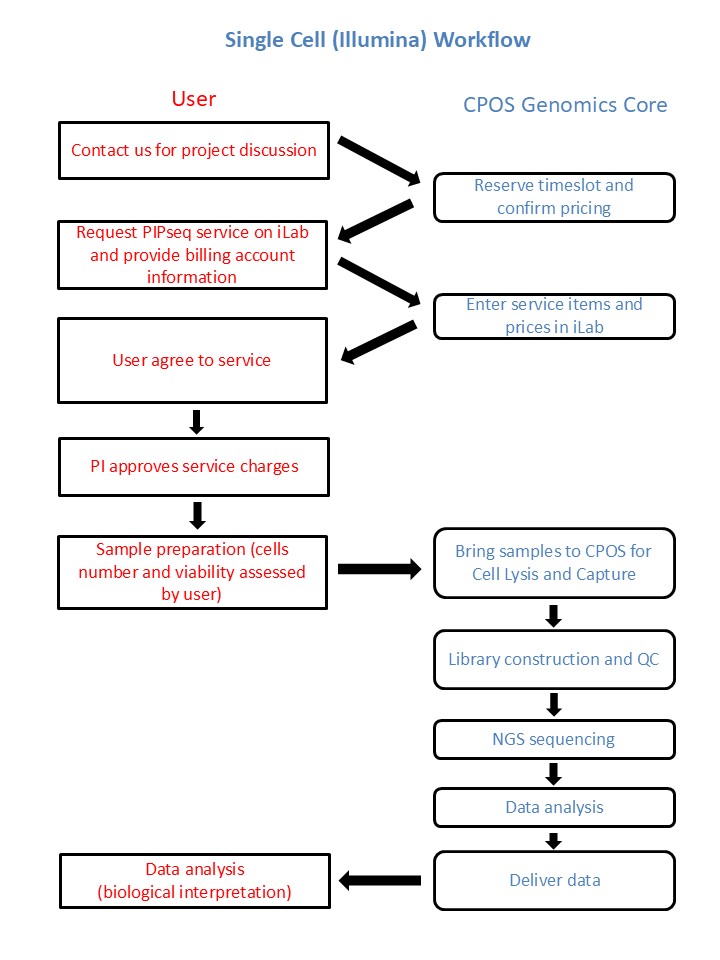
Workflow
Sample Requirement and Submission
Sample Requirement
Sample Type:
Single cell/nuclei suspension (Fresh / Fixed*)
Cell Size:
60 μm or smaller, well dissociated without clumps or doublets
Concentration:
3,400 cells/μl, minimum 50 μl
Viability:
>90%
Sample Buffer:
Cell Suspension Buffer (Illumina)**
* Fixed cells / nuclei need to be generated by DSP-Methanol Fixation. Alternative fixation approaches (e.g. formaldehyde) have not been validated and are not recommended.
**It is critical to complete the Cell Suspension Buffer wash of cell suspension prior to input into the PIPs to ensure any inhibitory reagents (e.g. FBS, BSA > 1%, high salt or calcium, carryover organic solvents) are sufficiently washed out of the cell suspension. The core will perform one wash step to change the buffer to Cell Suspension Buffer prior to mixing with PIPs.
Sample Submission
Due to project nature, samples need to be processed immediately upon submission. Please contact our colleagues in advance (via phone/email shown here) for timeslot reservation.
After sample submission date and details are confirmed, please submit service request through iLab. Please submit Single Cell (Illumina) sample submission form as attachment in iLab.
Terms of Service
The expected turnaround time for full service is usually 2-4 weeks from sample submission to sequencing data delivery. The turnaround time may vary depending on job demand.
Useful Links
Contact
Dr KWOK, Hin
Ms LI, Rachel




