Service Charges and Ordering
Visium HD Spatial Gene Expression
- Spatially profile transcriptome of tissue samples using probes targeting human or mouse transcriptomes
- 6.5 x 6.5 mm capture area with continuous lawn of spatially barcoded oligos
- Compatible with FFPE, fresh frozen or fixed frozen tissues
Service prices
The Core offers full service from sample QC, probe hybridization, probe capture using 10X Visium CytAssist instrument, subsequent library preparation, Illumina sequencing to data analysis by Space Ranger. Please contact Dr KWOK, Hin (singlecell10x.cpos@hku.hk / 2831-5483) for project discussion and service quotation.
Prices vary with the following factors:
- Size of target tissue region (affecting the sequencing cost)
- Tissue type (affecting the sequencing cost)
- Bioinformatics analysis needs
Service ordering
Please refer to Sample Requirement and Submission section for sample requirements.
Please submit service request through iLab.
Technical Details
Visium HD Slides
Each Visium HD slide contains two capture areas of 6.5 x 6.5 mm in size. In each capture area lies a continuous lawn of oligos comprised of 2 μm barcoded squares.
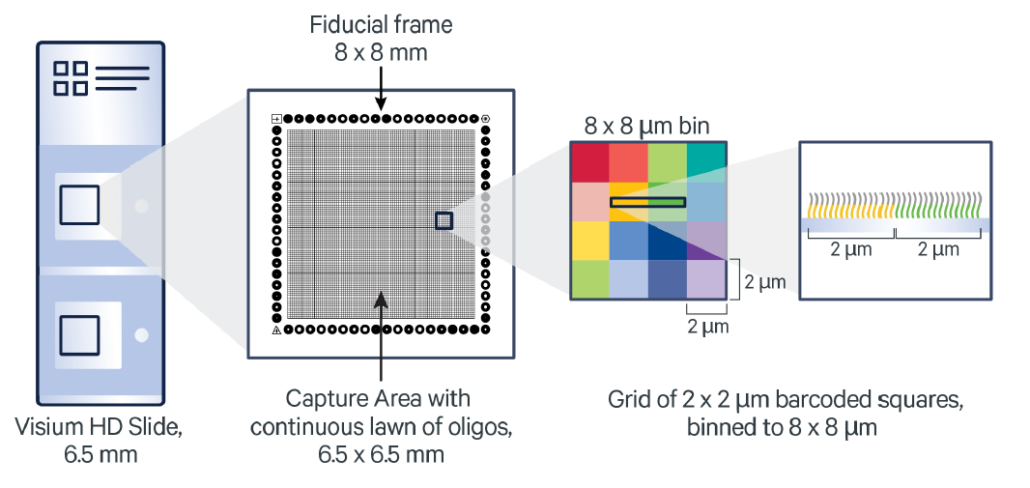
Source: 10X Genomics
Visium HD Gene Expression Library Preparation
In the Visium HD assay, tissue slides will first be stained and imaged. They will then be incubated with whole transcriptome probes to capture mRNA transcripts. After washing, probes will be released from the tissue slides and transferred to the Visium HD slide in the Visium CytAssist instrument. The instrument also captures a brightfield image during the run to record the spatial orientation. Probes hybridized to the Visium HD slide oligos will be used in library construction and downstream sequencing. Spatial barcodes are used to identify the original location of each transcript in the tissue.
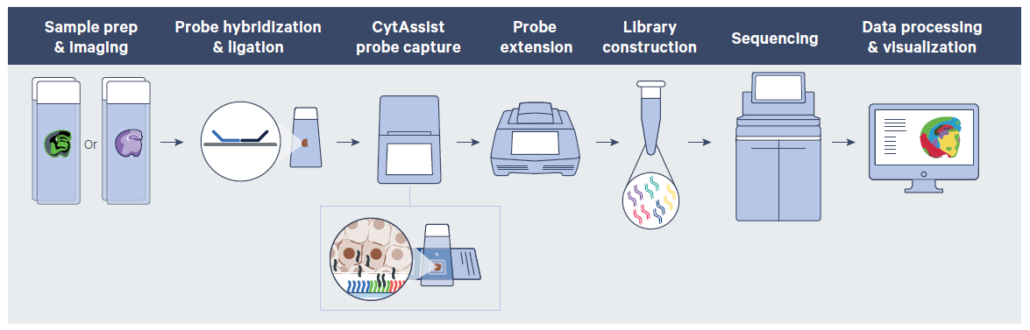
Source: 10X Genomics
Sequencing Run
Libraries generated are compatible with Illumina sequencers. In order to read the transcript sequences on one end, and the spatial barcode and UMI on the other end, paired-end sequencing reads are required.
Please refer to table below for vendor’s recommendation on the minimum sequencing depth required for each tissue type.
| X P | Visium HD | ||
| Tissue type | FFPE | Fresh frozen | Fixed frozen |
| No. of read pairs per capture area | 275 million | 700 million | 500 million |
| Run setting at CPOS | NovaSeq X Plus PE150bp |
Workflow
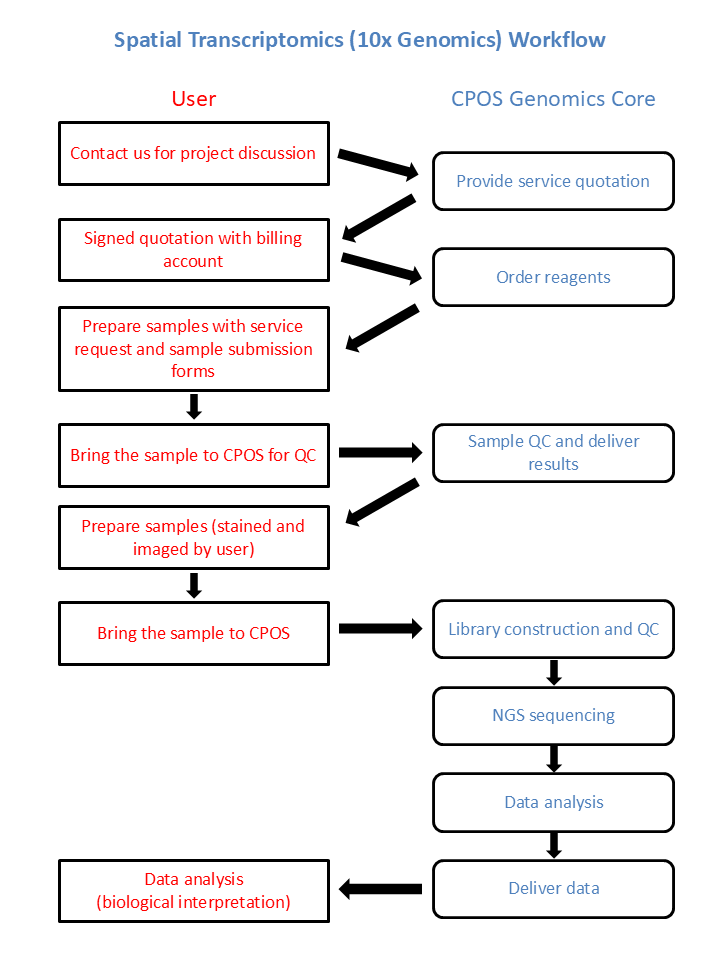
Sample Requirement and Submission
Sample Requirement
For Visium HD assays, users are required to submit samples for QC before actual experiment. Tissue sections for each sample should be placed into a 2 ml tube for RNA extraction and size QC. Users are welcome to submit more than 2 samples for QC.
For the actual experiment, users should place tissue sections on glass slides, perform staining and imaging. One tissue section should be placed on one glass slide. Users are recommended to prepare backup slides and select the best quality slides for downstream experiment based on imaging result. The same tissue section used for imaging should be submitted for the Visium experiment. Users must submit exactly 2 samples of the same species (human or mouse) per round.
Refer to below table for sample submission requirement.
| Purpose | Sample Type | FFPE | Fresh Frozen | Fixed Frozen |
| For Sample QC | Section Thickness | 10 μm | 25 μm | 25 μm |
| No. of Sections | 1-2 sections for ≥ 6.5 x 6.5 mm 4 sections for ≤ 6.5 x 6.5 mm | 4 sections (20-30 mg tissue) | 4 sections (20-30 mg tissue) | |
| Sample Format | Place tissue sections into 2 ml tubes | |||
| RNA QC Criteria | DV200 > 30% | RIN ≥ 4 | DV200 ≥ 50% | |
| Sample Submission Form for QC | Click here | |||
| For Visium HD | Section Thickness | 3-10 μm (Recommended: 5 μm) | 10-20 μm (Recommended: 10 μm) | |
| No. of Sections | 1 section per slide | |||
| Sample Format | Place tissue sections onto glass slides. Mount coverslip with glycerol. Click here for instructions on slide margins. | |||
| Sample Preparation Guide | Click here | Click here | Click here | |
| Imaging Guidelines | Click here | |||
| Tested Imaging System | Primo Micropatterning/Color Imaging System | |||
| Third-Party Items to Acquire | Click here | |||
| Sample Submission Form | Click here |
Sample Submission
Due to project nature, samples need to be processed immediately upon submission. Imaged slides must be submitted to Genomics Core no later than 5pm on working days. Please contact our colleagues in advance (via phone/email shown here) for timeslot reservation.
After sample submission date and details confirmed, please submit service request through iLab. Please submit sample submission form as attachment in iLab.
Terms of Service
CPOS cannot guarantee 100% accurate selection of region of interest on large tissues. We will in good faith produce the best output and quality data to users.
The expected turnaround time for sample QC is usually 1-2 weeks. For full service, it is usually 4-6 weeks from sample submission to sequencing data delivery. The turnaround time may vary depending on job demand.
Contact
Dr KWOK, Hin
Ms CHENG, Lilian




