Service Charges and Ordering
Single Cell 3′ Gene Expression Analysis
- 3′-end gene expression profiling of 500 – 10,000 cells per sample
Single Cell 5′ Gene Expression & Immune Profiling
- 5′-end gene expression profiling of 500 – 10,000 cells per sample
- Assemble, annotate full length T cell and/or B cell V(D)J sequences and call clonotypes
- Able to examine gene expression profile and resolve TCR/BCR repertoires from the same sample
Add-on Feature Barcode technology with Single Cell Gene Expression Assays
- Enable multiomic profiling by measuring cell surface protein expression or CRISPR perturbations simultaneously from the same cell
Single Cell Multiome ATAC + Gene Expression
- Multiomic profiling of the transcriptome (3’ gene expression) and open chromatin (ATAC-seq) from the same nucleus, for up to 10,000 nuclei per sample
Single Cell Gene Expression Flex
- Profile transcriptome of fixed samples using probes targeting human or mouse transcriptomes
- Compatible with cell suspensions, fresh or frozen tissues
- Multiplex up to 16 samples in one reaction
- Target up to 10,000 (4-plex reaction) or 8,000 (16-plex reaction) cells per sample
Add-on applications available for each assay:
| Single Cell 3’ Gene Expression Analysis | Single Cell 5′ Gene Expression & Immune Profiling | Single Cell Multiome ATAC + Gene Expression | Single Cell Gene Expression Flex | |
| V(D)J Sequencing | √ | |||
| Cell Surface Protein | √ | √ | Coming soon | |
| CRISPR Screening | √ | √ | ||
| Sample Multiplexing | √ |
Service prices
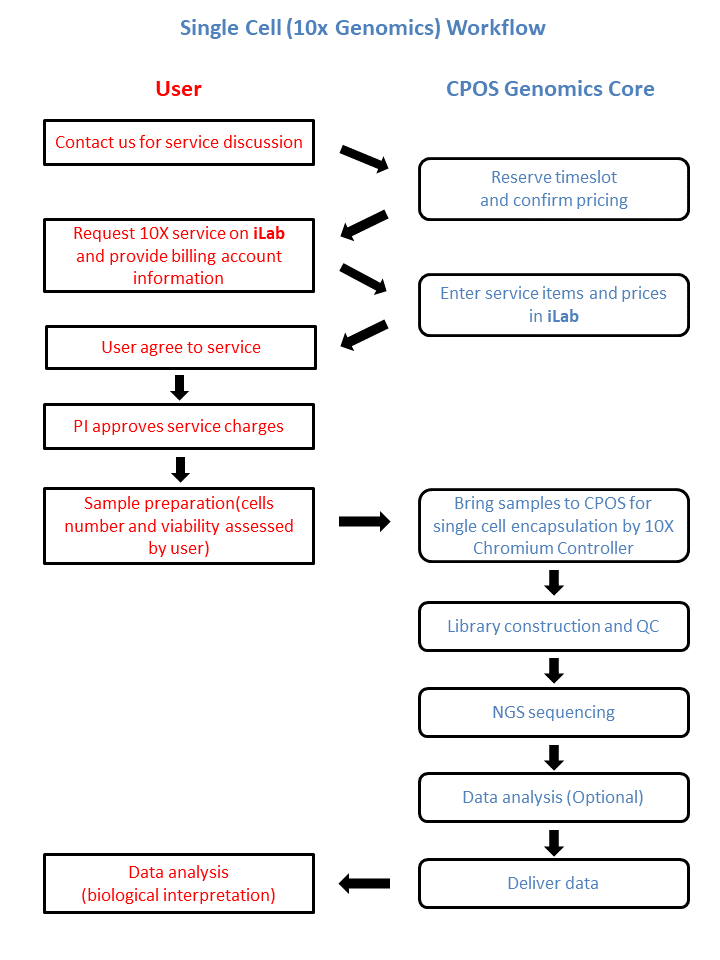
The Core offers full service from cell encapsulation using 10X Chromium iX, subsequent library preparation, Illumina sequencing to data analysis by Cell Ranger. Please contact Dr KWOK, Hin (singlecell10x.cpos@hku.hk / 2831-5483) for project discussion and service quotation.
Prices vary with the following factors:
- Number of samples per submission
- Applications (e.g. 3′ gene expression, 5′ gene expression, V(D)J analysis, etc.)
- Number of target cells (affecting the sequencing cost)
- Bioinformatics analysis needs
- Affiliated institute (HKU, local academics, overseas academics, etc.)
Service ordering
Please refer to Sample Requirement and Submission section for sample requirements.
Please submit service request through iLab.
Technical Details
Chromium Single Cell Encapsulation and Library Preparation
In 3′ or 5′ gene expression assay, cells will be encapsulated into Gel Beads-in-emulsion (GEMs) by the 10X Chromium iX, followed by reverse transcription and library preparation to become a pool of cDNA libraries. Transcripts from the same cell will be identified by a 10X barcode, while each of these transcripts will be discernible from one another by a Unique Molecular Identifier (UMI).
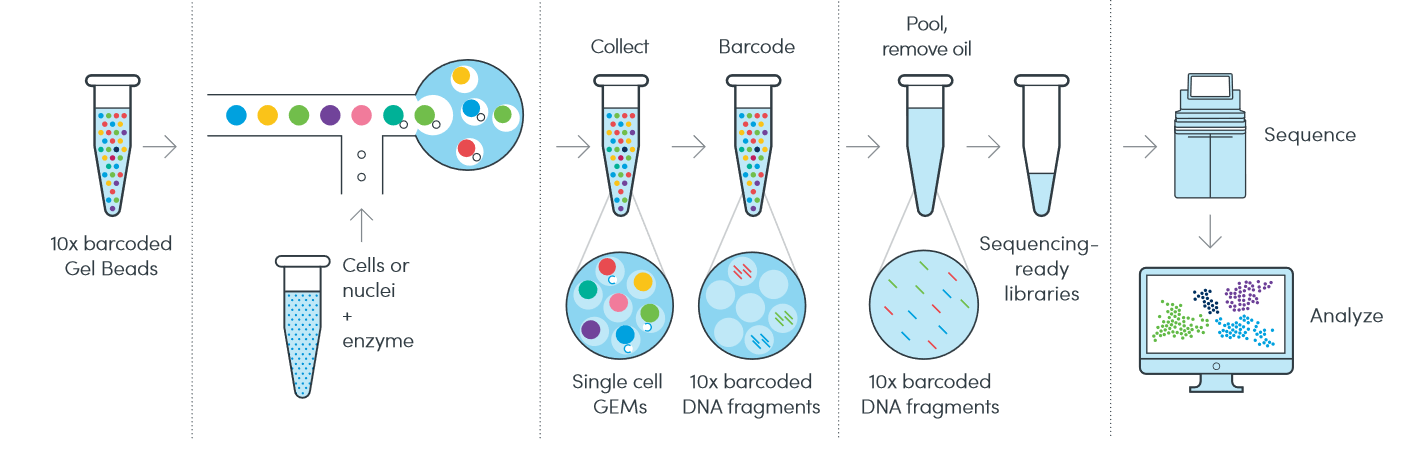
Source: 10X Genomics
For Multiome assay, nuclei will first undergo transposition in which transposase enters the nuclei and preferentially fragments the DNA in open regions of the chromatin. Transposed nuclei will be encapsulated following similar procedures described above to generate gene expression as well as ATAC libraries.
For Flex assay, samples will need to be fixed with formaldehyde and then incubated with whole transcriptome probes to capture mRNA transcripts. After hybridization, up to 16 samples can be pooled into one reaction for subsequent encapsulation and library preparation. On top of cell barcodes which are used to identify each single cell, probe barcodes are also present to assign cells to each sample.
Sequencing Run
Libraries generated are compatible with Illumina sequencers. In order to read the transcript sequences on one end, and the barcode and UMI on the other end, paired-end sequencing reads are required.
Please refer to table below for vendor’s recommendation on the minimum sequencing depth required for each application.
| Application | 3’, 5’ or Multiome Gene Expression Library | V(D)J Library | Feature Barcode Library | Multiome ATAC Library | Gene Expression Flex Library |
| No. of read pairs per cell/nucleus | 20,000 | 5,000 | 5,000 | 25,000 | 10,000 |
| Run setting at CPOS | NovaSeq 6000 PE150bp | NovaSeq 6000 PE150bp | NovaSeq 6000 PE150bp | NovaSeq 6000 PE50bp (Due to custom run cycles, users are required to commit their own sequencing run) | NovaSeq 6000 PE150bp |
Workflow
Sample Requirement and Submission
Sample Requirement
Refer to below table for sample submission requirement of each application.
| Application | 3’ or 5' Gene Expression | Multiome ATAC + Gene Expression | Gene Expression Flex |
| Sample Type | Single cell/nuclei suspension | Single nuclei suspension | Fixed cell/nuclei suspension |
| Cell Size | 30 μm or smaller, well dissociated without clumps or doublets | ||
| Concentration | 500 – 2,000 cells/μl, minimum 50 μl | 3,000 – 8,000 nuclei/μl, minimum 10 μl | No restriction on concentration. Please prepare >100,000 fixed cells/nuclei. |
| Viability | Cells: ≥85% Nuclei: <5% | <5% | N/A |
| Sample Buffer | 1x PBS + 0.04% BSA (or other buffers recommended by vendor) Avoid excessive amounts of EDTA (>0.1mM) or magnesium (>3mM), and any surfactants (Tween-20, etc.) | Diluted Nuclei Buffer with RNase inhibitor Stock solution of Nuclei Buffer (10X Genomics) will be provided by CPOS. | Quenching Buffer (10X Genomics) or Tissue Resuspension Buffer |
| Sample Preparation Guide | Click here | Click here | Click here |
| Sample Submission Form | Click here | Click here | Click here |
Sample Submission
Due to project nature, samples need to be processed immediately upon submission. Please contact our colleagues in advance (via phone/email shown here) for timeslot reservation.
After sample submission date and details confirmed, please submit service request through iLab. Please submit Single Cell (10X Genomics) sample submission form as attachment in iLab.
Terms of Service
Due to technology limitation, there may be occasional wetting failure or clogging during encapsulation, leading to unusable reaction. CPOS cannot ensure successful encapsulation for every sample. We will in good faith produce the best output and quality data to users. In case of failed experiment, we will contact users immediately to discuss how to proceed.
The expected turnaround time for full service is usually 2-4 weeks from sample submission to sequencing data delivery. The turnaround time may vary depending on job demand.
Contact
Dr KWOK, Hin
Ms CHENG, Lilian




