Bioinformatics Core
If you received an error of “Authentication failed; Critical error: Could not connect to server” during connection to sFTP server.
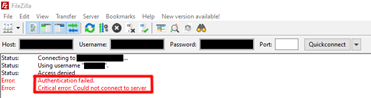 This is likely due to username or password may contain some “look-alike” characters, such as “I”, “1”, “0” or “O”. You are suggested to copy and paste the information for connection again.
This is likely due to username or password may contain some “look-alike” characters, such as “I”, “1”, “0” or “O”. You are suggested to copy and paste the information for connection again.
If you encountered an error of “ssh_init: Host does not exist; Could not connect to server” during connection to sFTP server.
The error may be caused by your network blocking our sFTP server or unstable network. Please try another network for connection again. Also, due to instability of Wi-Fi, you are highly recommended to download the files with a more stable network connection, such as wired LAN.
If you received an error of “error while writing local files” when downloading the files from sFTP server.
The error may be caused by insufficient disk space in your computer or unstable network. Please check if your hard disk has sufficient free space for storing the data. Also, due to instability of Wi-Fi, you are highly recommended to download the files with a more stable network connection, such as wired LAN.
If you are unable to extract the second zip file (zip.002) with provided password.
The zip.001 and zip.002 are the split zip files of the original folder. Only zip.001 needs to be extracted and the remaining zip files (zip.002 and/or zip.003 if any) will be unzipped automatically into a single folder. Hence, please do not unzip other zip files except zip.001.
If you encountered an error of “Cannot open the file as zip archive” during extraction.
![Cannot open the file as [zip] archive](../wp-content/uploads/2021/06/BF_FAQ_DataExtractionQ2.png)
The files may be corrupted due to unsuccessful downloading from our sFTP server. Please ensure that you have followed the instruction to download the files and verify the file integrity with WinMD5Free.
If you are unable to extract due to “Wrong password” during extraction.

This is likely due to username or password may contain some “look-alike” characters, such as “I”, “1”, “0” or “O”. You are suggested to copy and paste the password for extraction again.
If your extracted files contain no data (0KB).
The files may not have extracted successfully due to wrong unzip password. You are suggested to copy and paste the password for extraction again.
If you received other error messages during file extraction.
Using our recommended extraction software (7zip) will generally solve the error. Please contact us if the error still persists after using 7zip.
If you want to understand the deliverables provided by CPOS bioinformatics analysis service.
For standard analysis, please refer to the deliverable table of different project types. If you would like to have additional deliverables, please contact us for a discussion.
If you wish to find out about the turnaround time of a standard bioinformatics analysis.
The turnaround time for a standard bioinformatics analysis is around 2 weeks, upon the raw sequencing data is ready and required information (e.g. sample species, comparison list) is received beforehand.
If you want to know whether your sequencing data contains adapter sequences.
In general, the sequencing data contains adapter sequences. The adapters will only be trimmed upon request in the MiSeq sequencing service.
If you need adapter information for your sequencing data.
If you want to trim the adapters for your sequencing data, you may refer to the first pair of adapter sequences in this website.
Read 1: AGATCGGAAGAGCACACGTCTGAACTCCAGTCA
Read 2: AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT
Please remember to check the adapter-trimmed fastq files before the downstream analysis.
If you are going to analyze the Partek project provided from July 2021 onwards, in Partek Genomics Suite.
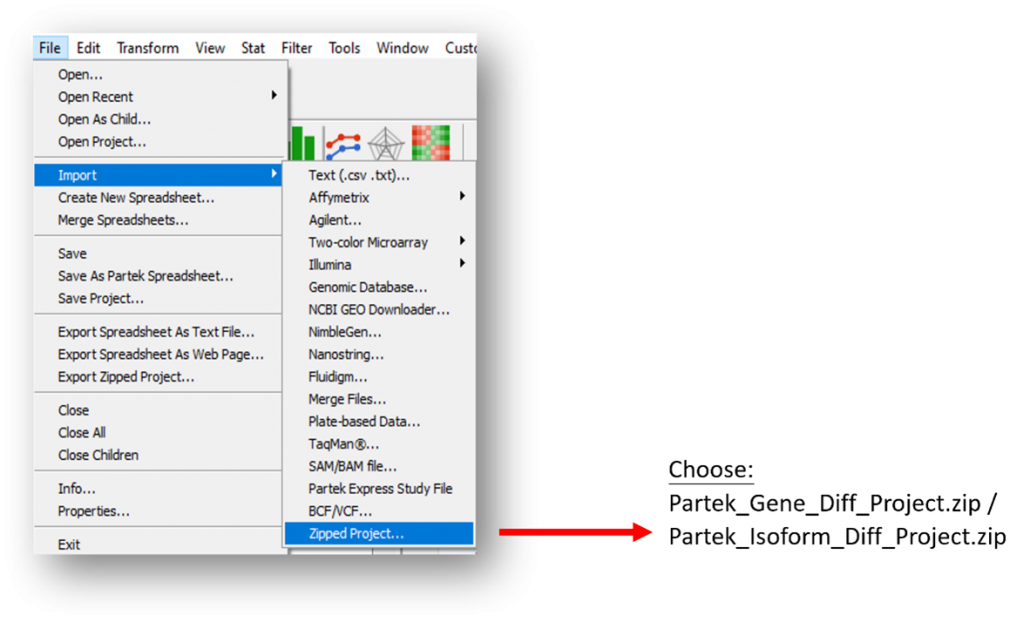
The file format of Partek project provided in RNA-Seq analysis will be changed from “.ppj” to “.zip” from July 2021.
To import the project, click “File” -> “Import” -> “Zipped Project…” -> Choose the project to be imported
**Other operations in PGS remain unchanged.
If you are asked to provide “Probeset Annotation” during pathway analysis in Partek Genomics Suite.

To specify, click “Browse” -> Choose the corresponding text file of your comparison
(e.g. AvsB_Partek_Gene_Diff.txt / AvsB_Partek_Isoform_Diff.txt) -> Click “OK”.
Proteomics and Metabolomics Core
What is the sequence coverage obtained in a typical protein identification?
Generally, the sequence coverage is between 10% and 90% per protein, which is dependent on the protein abundance and protein nature.
What are the most common reasons for obtaining poor mass spectrometric data?
How do I avoid contamination with keratin?
When can I expect my results back?
The typical turn around time is 2 weeks for simple Protein Profiling/PTM IDs, routine services for targeted metabolite quantification (GC-MS/MS, and LC-MS/MS); 3 weeks for Protein ID & Quant (Label-free Protein Quantification), 4 Weeks for Label-based (TMT/iTRAQ) protein ID & Quantification months, and 6 weeks for Label based phosphoproteomics services.
Biobank
WHO can use the Faculty Biobank?
Staff within the HKU Faculty of Medicine are eligible and have priority to use the Biobank services. Other HKU staff may enquire through email: biobank.cpos@hku.hk. Long-term freezer storage is only for Staff within the HKU Faculty of Medicine.
WHAT types of services does the Biobank provide?
The Biobank provides sample storage services and some sample processing services, such as separation of plasma/serum/buffy coat from whole blood, sample aliquot services etc. Please email biobank.cpos@hku.hk to discuss your needs.
HOW is the Biobank managed?
The Biobank is established by the LKS Faculty of Medicine as part of its Core Facilities. The day-to-day operation and management of the Biobank is handled by the Centre for PanorOmic Sciences (CPOS) – Biobank Core staff.
Is there any RESTRICTION on what can be stored in the Biobank?
-
- WHO Risk Group 3 and 4 microorganisms cultured in the laboratory, e.g. Influenza virus strains such as H5N1 and H7N9, Mycobacterium tuberculosis, Ebola, Marburg, Herpesvirus simiae etc.
- High titre stocks of Risk Group 2 microorganisms of known significant risk, e.g. Burkholderia pseudomallei, Bordetella pertussis, Corynebacterium diphtheria, Neisseria meningitides etc.
- Clinical or environmental samples that have been confirmed to carry the above mentioned microorganisms.
- Radioactive materials
A declaration form with additional details needs to be signed by each P.I. and user prior to using the Biobank.
HOW MUCH does it cost?
The Faculty is providing subsidy for the usage of Biobank, while some operating cost is to be recovered based on resources consumed. You may find some standard charges on the “Service Charges” page. For other services, please email biobank.cpos@hku.hk for discussions and quotations.
How do I GET STARTED if I wish to use the Biobank?
Please go through the information on this website, then email biobank.cpos@hku.hk
Information to facilitate effective follow-up may include:
- Your name, department and phone number
- P.I. name, department and email
- Samples nature, e.g. whole blood, buffy coat, plasma, serum, DNA, RNA, virus, bacteria
- Sample size, source and pathogenicity involved
- Storage condition required
- For sample storage, current box/vessel types and sizes
- Other specific needs
Imaging and Flow Cytometry Core
How to use the PPMS as a user?
Please refer to the user’s guide Here.
How to use the PPMS as a supervisor?
Please refer to the supervisor’s guide Here
How to register for access to Imaging and Flow Cytometry Core Data Transfer Server?
- Register in the user registration page http://ifcs.cpos.hku.hk/share-drive/create_user.php
with any Internet browser.- Fill in the E-mail with HKU email Only (uid@hku.hk or uid@connect.hku.hk), Do NOT use email alias. Fill in your full name and password
- Click ‘Register’ to submit and generate a confirmation email which will be sent to your HKU E-mail account.
- Check your registered mailbox for email titled “CPOS Imaging and Flow Cytometry Core Network Server Account Registration Confirmation”.
- Complete the registration by clicking on the confirmation link in the email received.
How to upload data to Imaging and Flow Cytometry Core Server?
How to download data from Imaging and Flow Cytometry Core Server?
Imaging and Flow Cytometry Core Server Connection and Data Downloading Instruction – Windows 10.
Imaging and Flow Cytometry Core Server Connection and Data Downloading Instruction – Windows 7.
Imaging and Flow Cytometry Core Server Connection and Data Downloading Instruction – Mac.
Imaging and Flow Cytometry Core Server Connection and Data Downloading Instruction– Linux.
Imaging and Flow Cytometry Core SOP – Accessing DTS with Updated Credentials on macOS.
For downloading dara from non-HKU network, please connect to HKUVPN prior to mapping the drive in server.
How to reset password for Data Transfer Server of Imaging and Flow Cytometry Core?
Steps to reset password
- Use an Internet browser to open the password reset page at http://ifcs.cpos.hku.hk/share-drive/passwd.php
- Enter your HKU email address
- Check the link in the password reset email sent to your HKU mailbox and enter your new password in the page to reset your password
Bioreagent Core
How do I place order in the Online Ordering System?
Please follow the Staff User guide here
What items are being stocked in the core?
There are >20 category of common consumables and reagents from >60 well-known brands, Including lab consumables for cell culture, microbiology, imaging & flow cytometry, molecular as well as lab routine operation. Please refer to the Bioreagent Core Online Ordering System (BROS) for the detailed item list.
Can new items to be added in BR Core?
New item suggestions are welcome. Please provide the desired product details, consummation rate, recent price to contact bioreagent.cpos@hku.hk for our consideration.
Where does the delivery service cover?
The delivery service serves orders from labs in Queen Mary Hospital, Sassoon Road campus including Laboratory Block, JCBIR, CCMR, Chinese Medicine Building. For lab in other location, please arrange self pick up from the Core warehouse within 5 working days from the order confirmed date.
When can I receive my products after I have confirmed my order?
For orders for Laboratory Block, Items in confirmed orders will be delivered within 3 working days. Schedule is subject to change depends on the actual situation. For orders from QMH, JCBIR and buildings along Sassoon Road, weekly delivery will be offer on the fixed date as schedule shown in online ordering system login message. For urgent pick up, please contact us via phone or email.
When the items can be collected by self pick up?
Item can be Self pick ups for urgent situation. Service available time is 9:30-12:00; 14:00-17:00. Please notice us via phone or email in advance to arrange self pick up. Walk-in self pick up requests will not be entertained.
What procedure is needed for Core’s Delivery Note (DN) and Monthly Invoice
There are two type of Delivery Notes issued by Bioreagent Core: Consignment Delivery Note and Delivery Note.
Consignment Delivery Note are issued for items for consignment that are distributed by Bioreagent Core. The corresponding vendors will issue invoices to the requester in due course. Upon receive the invoice from vendors, the invoice should be proceeded as direct invoice for the purchase. Payment should be initiated by submitting the endorsed invoice to your department and then HKU FEO.
Delivery Note are issued for items that are retailed by Bioreagent Core. At the beginning of each month, a Monthly Invoice will be issued to group supervisors for all orders collected by the end of the previous month. The charges will be settled by interdepartmental transfer from the funding account provided during order. Payment would be initiated by Core’s submission of the monthly consolidated amount to HKU FEO. Therefore please DO NOT submit the invoice to HKU FEO.
How to place order with total amount exceeds approval limit. What should I do after I have submitted the order to my supervisor?
Order with amount exceeding the approval limit should be submitted for supervisor’s approval. To get the order approved, you may either 1) attach in the order an endorsed printing of corresponding IR, or 2) invite your Supervisor to approve the order in the online system by submitting the order to your supervisor, 3) write email to your supervisor for approval of the purchase with carbon copy (cc) to bioreaeagent.cpos@hku.hk.
How to change payment account in an invoice?
To amend the Charging Account(s) in an invoice, please send an email to enquiry.cpos@hku.hk within 30 days from the date of the invoice, including the information of 1) Invoice No. 2) Group Name 3) Item Request No. 4) New Charging Account No.
How can order and expense of each group be reviewed?
Group Transection Report can be generated on the system for the order, items’ details, account details, unit price, total amount etc. To generate the report, click ‘Group Transection Report’ under ‘Report’ and select appropriate Date, Vendor and Group.
Bioresearch Support Core
How to use the PPMS as a user?
Please refer to the user’s guide Here.
How to use the PPMS as a supervisor?
Please refer to the supervisor’s guide Here
How to register for access to the Core Data Transfer Server?
1. Register in the user registration page
http://ifcs.cpos.hku.hk/share-drive/create_user.php with any Internet browser.
- Fill in the E-mail with HKU email Only (uid@hku.hk or uid@connect.hku.hk), Do NOT use email alias. Fill in your full name and password
- Click ‘Register’ to submit and generate a confirmation email which will be sent to your HKU E-mail account.
- Check your registered mailbox for email titled “CPOS Network Server Account Registration Confirmation”.
- Complete the registration by clicking on the confirmation link in the email received
How to upload data to the Core Server?
How to download data from the Core Server?
Server Connection and Data Downloading Instruction – Windows 10.
Server Connection and Data Downloading Instruction – Mac.
Bioresearch Support Core SOP – Accessing DTS with Updated Credentials on macOS.
For downloading dara from non-HKU network, please connect to HKUVPN prior to mapping the drive in server.
How to reset password for Data Transfer Server of the Core?
Steps to reset password
- Use an Internet browser to open the password reset page at http://ifcs.cpos.hku.hk/share-drive/passwd.php
- Enter your HKU email address
- Check the link in the password reset email sent to your HKU mailbox and enter your new password in the page to reset your password

